Introduction: Phenothiazine 1 also known as thiodiphenyl amine is a heterocyclic ring system containing 1, 4-thiazine moiety bounded by two benzene rings in an ortho-fused pattern 1. This linear parent compound 1 and its derivatives have been known to exhibit promising medicinal properties2. Other nonlinear (angular) phenothiazine derivatives3 of the types 2 and 3 have also be reported. They possess very useful properties4 and have been used in the production of pesticides5, dyes and pigments5, antioxidants6, drugs7 and many other pharmaceutical products8
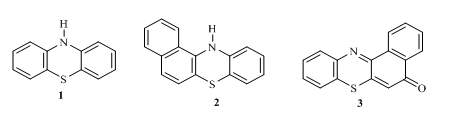
Variation of substituents and modification made on the parent phenothiazinehas given birth to a myriad of derivatives of interest with different range applications 9-11. For instance chlorproma-zine, an antihistamine could reduce psychotic symptoms 12 while promethazine is as an anesthetic agents 13.
Sulphonamide 4 also known as p– aminobenzenesulphonamide and it derivatives of the type 5 were among the first set of antibiotics used to prevent and treat different disease 14.
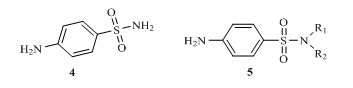
R1/R2 = H, alkyl, aryl or hetero aryl group
Figure 2: Structures of p– aminobenzenesulphonamide and its derivatives
As a result of structural modifications on the parent sulphonamide compound 4, many derivatives called sulpha drugs are now available 15. There are over thirty (30) drugs with this functionality which have been used to handle different conditions, for instance sulphathiazole 6 is used to treat infections, sulphapyridine 7 used for treating pneumonia, sulphaxozole 8 has been used fight bacteria as well as treat protozoan and fungi infections 16. Others like cetazolamide (carbonic anhydrase inhibitor) 9 have been used to reduce inflammation and as inhibitors of certain receptors in the body 17.
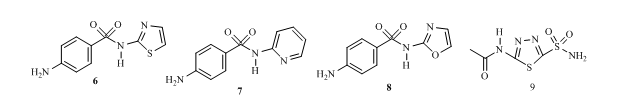
Fig 2: Examples of sulphonamide drugs
Studies have shown that combinatorial method of drug synthesis is a recent development used in pharmaceutical industries to synthesize drug molecules 18. This is to tackle the resistance that microorganisms have developed against many antibiotics. Therefore in order to overcome such resistance different compounds are now fused to form single molecules to offer better activities via synergy. Phenothiazine and sulphonamide individually have been known to exhibit very good biological activities, however, there are few previous reports available on phenothiazine and sulphonamide functionalities fused into single molecules. Most importantly, studies on these new compound known as N-(7-nitro-10H-phenothiazin-3-yl)benzenesulphonamide derivatives have not carried out. In this work we reported the synthesis of compounds bearing the above functionalities in single molecules with better biological activities.
2.0 Materials and methods
2.1 General
All reagents were obtained from Aldrich and were used as received. 1H-NMR and 13C-NMR spectra were run on Joel 400MHz spectrometers in CDCl3 using TMS as internal threshold. FTIR Spectroscopy of the compounds were run in PerkinElmer Spectrum version 10.03.06 and the bands given in wave number. The mass spectroscopy was carried out using micro time of flight electrospray mass spectrometer (Aerodyn Research Inc USA). Melting points were determined in open capillary tubes and are uncorrected.
- Synthesis of N1-(4-nitrophenyl)benzene-1,4-diamine 12
A combination of 4-nitroaniline 10 (23.0 mmol) and 4-chloroaniline 11 (23.0 mmol) in dimethylformamide (10 ml), triethylamine (3 ml) and n-butanol (100 ml) was swirled under reflux for 6h in an oil bath and checked via TLC screening. At the expiration of the process, the mixture was filtered and poured into a beaker containing crushed ice and acidified with 2 M HCl (30.0 ml) to precipitate the product which was birthed with distilled water (3×50 ml) and vacuum dried. The dried product was recrystallized from methanol to give N1-(4-nitrophenyl) benzene-1, 4-diamine 12, characterized thus,
Yield 75.7%, melting point 134-135oC. FTIR (KBr, cm-1): 3354.82 (N-H stretch), 3217.95, (NH2 1o amine), 3114.07 (C-H aromatic), 1619.34 (C=N, C=N). 1H NMR (δ): 8.36 (s, 1H, NH amide), 8.12-8.03 (q 2H, 4.4 Hz, ArH), 7.84-7.72 (q, 2H, 6.1 Hz, ArH), 7.53-7.47 (q, 2H, 3.2 Hz, ArH), 6.81-6.80 (q, 2H, 0.1 Hz, ArH), 4.50 (s, 2H, NH2). 13C NMR (δ): 148.06, 142.65, 132.86, 129.55, 129.44, 128.58, 127.82, 127.01, 125.22, 125.102, 125.09, 118.47 (12 aromatic carbons). HRMS-ESI (m/z) for C12H11N3O2: found 232.09 [M– 3H]+, calculated 229.24.
2.1.3 Synthesis of 7-nitro-10H-phenothiazin-3-amine 13
A mixture of N1-(4-nitrophenyl) benzene-1, 4-diamine 12 (11 mmol), sulphur powder (19 mmol) and iodine crystals (0.8 mmol) in n-butanol (100 ml) was agitated under reflux in an oil bath for 7h and checked via TLC screening. At the termination of the process, the mixture was cooled in an ice bath, filtered and dried. The product was added into a beaker containing n-butanol (50 mL) and activated charcoal and heated until solid dissolved. The solution was then filtered hot, cooled in ice bath to obtain yellow crystals of 7-nitro-10H-phenothiazin-3-amine 13. The compound was characterized as thus;
Yield 90.0%, melting point 109-110oC. FTIR (KBr, cm-1): 3427.10 (N-H stretch), 3121.25, (NH2, 1o amine), 2996.13 (C-H aromatic), 1633.91 (C=N, C=C). 1H NMR (δ): 7.72 (s, 1H, ArH), 7.70 (s, 1H, ArH), 7.49-7.26 (q, 2H, 10.6 Hz, ArH), 7.24 (s, 1H, ArH), 6.40 (s, 1H, ArH), 4.72 (s, 2H, NH2). 13C NMR (δ): 148.06, 142.64, 132.64, 129.55, 129.44, 128.30, 127.02, 127.01, 125.22, 125.18, 125.09, 118.47 (12 aromatic carbons). HRMS-ESI (m/z) forC12H9N3O2 S: found 232 [M+H] +, calculated 259.28. (12 aromatic carbons).
2.1.4 Synthesis of 7-nitro-10H-phenothiazin-3-benzenesulphonamide derivatives 15 a-c
A combination of 7-nitro-10H-phenothiazin-3-amine 14 (5.8 mol) and triethyl amine (3 mL) in n-butanol (100 ml) and DMF (5mL) was poured into 100 mL flask and agitated via a magnetic stirrer until all the solutes dissolved. Thereafter, substituted arylsulphonyl chlorides 15 a-c (9.4 mol) were added in portions over a duration of 1 h. The bulk of the flask was refluxed for 7h in an oil bath and checked via TLC screening. At the expiration of the process, the pH was adjusted from 7 to 2 with 2 M HCl (5 mL). The organic layer was retrieved and birthed with brine (3 x 50 mL) via a separating and vacuum dried to give final products 15 a-c. The compounds were then characterized as follows:
N-(7-Nitro-10H-phenothiazin-3-yl)benzenesulphonamide 15a
Yield 82.5%, melting point 800C-81oC. FTIR (KBr, cm-1): 3365.97 (N-H stretch), 3121.78, (C-H aromatic) 1625.622cm-1 (C=N, C=C), and 1341.25 (S=O vibration of sulphonamide). 1H NMR (δ): 8.36 (s, 1H, NH amide), 8.07 (s, 1H, ArH), 7.86 -7.84 (q, 2H, 1.0 Hz, ArH), 7.720 (s, 1H, ArH), 7.53-7.47 (q, 2H, 3.2 Hz, ArH), 7.46 (s, 1H, ArH), 6.81 (s, 1H, ArH), 6.38 (s,1H, ArH). 13C NMR (δ): 148.06, 145.01, 142.50, 140.00, 132.99, 129.55, 129.54, 129.44, 128.58, 129.51, 127.82, 127.21, 127.01, 125.22, 125.10, 125.09, 122.919, 118.47 (18 aromatic carbons). HRMS-ESI (m/z) for C18H13N3O4 S2: found 399 [M]+, calculated 399.44.
4-Methyl-N-(7-nitro-10H-phenothiazin-3-yl) benzenesulphonamide 15b
Yield 80.6%, melting point 141oC-142oC. FTIR (KBr, cm-1): 3229.11 (N-H stretch), 3075.25 (C-H aromatic), 1626.05 (C=N, C=C stretch), 1385.04 (S=O vibration of sulphonamide) and 1H NMR (δ): 9.00(s,1H, NH), 8.37(s,1H, ArH), 8.07-8.05 (q, 2H, 0.9 Hz, ArH), 7.83 (s, 1H, ArH), 7.781 (s, 1H, ArH), 7.780 (s, 1H, ArH), 7.501(s,1H, ArH), 7.500 (s,1H, ArH), 2.730 (s ,3H, CH3). 13C NMR (δ): 148.06, 145.01, 143.00, 140.00, 133.00, 129.33, 129.50, 129.44, 128.58, 127.82, 127.82, 127.01, 127.013, 125.22, 125.10, 125.09, 122.00, 110.47, 23.85 (18 aromatic carbons). HRMS-ESI (m/z) for C19H15N3O4 S2: found 413 [M] +, calculated 413.47.
4-Nitro-N-(7-nitro-10H-phenothiazin-3-yl) benzenesulphonamide 15c
Yield 81.9%, melting point 1000C-101oC. FTIR (KBr, cm-1): 3153.152 (N-H stretch), 3047.637 (C-H aromatic), 1625.611 (C=N C=N stretch), 1341.258 (S=O vibration of sulphonamide). 1H NMR (δ): 9.00 (s, 1H, NH), 7.72-7.70 (q, 2H, 1.0 Hz, ArH), 7.33 (s, 2H, ArH), 7.32 (s,1H, ArH), 7.317 (s,1H, ArH), 7.00 (s, 1H, ArH), 6.79 (s, 1H, ArH), 6.65 (s,1H, ArH), 6.40 (s,1H, ArH). 13C NMR (δ): 151.26, 148.06, 143.01, 140.00, 133.01, 129.55, 129.44, 129.35, 128.58, 127.82, 127.01, 125.22, 125.10, 125.09, 122.42, 121.43, 118.47, 111.63 (18 aromatic carbons). HRMS-ESI (m/z) for C18H12N4O6 S2: found 444 [M] +, calculated 444.44.
2.1.5 In silico Studies
Molecular docking was carried out to predict interactions of the protein targets (7R1M, 7AB4, 2YXB and 5ZVP) for Staphylococcus aureus, bacillus cereus, Escherichia coli, Salmonella, Aspergillus fumigatus respectively with the synthesized compounds. Ciprofloxacin (antibacterial) and Ketoconazole (antifungal) were used as control ligands. The docking study was done using AutoDock tools GUI (graphical user interface) (version 4.2) and was based on the Vina script. Their 3D crystal structures obtained from protein data bank thus; (PDB: 7D8I with 1.62Å resolution), (PDB: 7R1M with 1.64Å resolution), (PDB: 7AB4 with 3.34Å resolution), (PDB: 4YXB with 2.56Å resolution), (PDB: 5ZVP with 1.42Å resolution) 19-21. The structures were optimized via Gaussian 09 while the grid box sizes were set via AutoDock tool with dimensions, X= 24 Y= 24 Z= 24 and 1.00 Å as the grid spacing. Lamarckian genetic algorithm protocol was employed to generate the optimum binding site for the ligand and Gasteiger charges were added via the AutoDock Tool’s graphical user interface supplied by MGL Tools 21, 22.
- Drug likeness of prepared compounds
The drug likeness properties of the derivatives was estimated via SwissADME online tool.
- Biological studies
The antimicrobial screening for substituted the N-(7-nitro-10H-phenothiazin-3-yl)benzene- sulphonamide derivatives was done via the agar well diffusion method following the National Committee for Clinical Laboratory Standards NCCLS guidelines (2002).
- Preparation of Media
Mannitol salt agar (11.0 g) was used to prepare the culture medium for staphylococcus aureus strain, Salmonella-shigella agar (60.0 g) for salmonella and potato dextrose agar (39.0 g) for aspergillus fumigatus. Distilled water (1000 mL) was added to the media contained in three separate conical flasks. The flasks were sealed with wool and foil and autoclaved for 15mins at 1210C. The cultured agar media were then poured into four sterilized petri dishes and allowed to solidify.
- Culturing of the isolates
Inoculating loops were placed in Bunsen burner blue flame for sterilization, removed and allowed to cool. The cultured media in the various petri dishes were streaked using the loop and incubated for 24 hours at 25°C.
2.2.0 Nutrient agar preparation
Nutrient agar powder (28.0g) was added to distilled water (1000 mL) in a conical flask and swirled to ensure complete dissolution. The flask was autoclaved at 121°C for 15 minutes, cooled at 37°C and was then poured into the petri dishes and allowed to solidify.
2.2.1 Preparation of selected samples
7-Nitro-10H-phenothiazin-3-amine 13, N-(7-nitro-10H-phenothiazin-3-yl)benzenesulphonamide 15a, 4-methyl-N-(7-nitro-10H-phenothiazin-3-yl)benzenesulphonamide 15b and 4-nitro-N-(7-nitro-10H-phenothiazin-3-yl)benzenesulph-onamide 15c, each was prepared in four different concentrations (2.5 mg/mL, 5 mg/mL, 1.25 mg/mL, 0.625 mg/mL) using dimethyl sulfoxide (DMSO) as solvent and poured into four test tubes containing DMSO (2 mL) and labelled accordingly. 1mL of each of the above samples was added to each of the petri dishes containing the cultured microorganisms and their zones of inhibition were measured.
- Determination of antimicrobial activity
Sterilized cock borers were used to five (5) holes in each of the petri dishes containing the cultured media of the microorganisms and labelled accordingly. 1 mL of each of the concentrations of the above compounds and control (DMSO) was added into the holes and left stand at room temperature for 48 hours. Thereafter, their zones of inhibitions were determined.
3.0 Results and Discussions
3.1 Chemistry
The base catalyzed reaction of 4-nitroaniline 10 and 4-chloroaniline 11 in the presence of n- butanol, triethylamine and dimethylformamide (DMF) gave 7-nitro-10H-phenothiazin-3-amine 12 which was further treated withsulphur powder and catalytic amount of iodine crystals using the same reaction conditions to give 7-nitro-10H-phenothiazine-3-amine 13. Subsequent treatment of compound 13 with three appropriate arylsulphonyl chlorides 14 a-c in the presence of triethyl amine and n-butanol yielded the corresponding substituted 7-nitro-10H-phenothiazin-3yl-sulphonamide derivatives 15 a-c as shown in scheme 1 below.
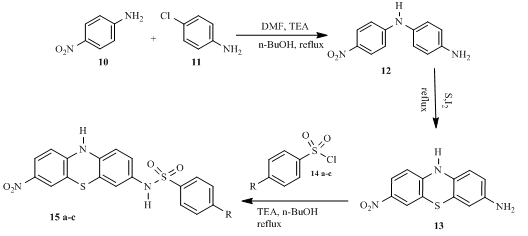
Scheme 1: Synthetic route of 7-nitro-10H-phenothiazin-3yl-sulphonamide derivatives
In the FTIR spectrum of 1-(4-nitrophenyl)benzene-1,4-diamine 12, the band were observed at the following regions 3354.82 cm-1, 3217.95, 3114.07 cm-1 and 1619.342 cm-1 corresponding to NH, NH2 C=N and C=C stretch respectively. In the 1H-NMR spectrum, one peak was found at δ 8.36 is for an amide proton, the peaks were also found at δ 8.126-8.03 – 6.810 representing seven aromatic protons while the peak at δ 4.50 is for the two proton of amine functionality. In the 13C NMR spectrum, the peaks at δ148.06 -118.47 are for the twelve aromatic carbons present in the compound. The molecular ion [M+] has a peak at 232.09 in the mass spectrum. For compound 13, the FTIR spectrum the band at 3427.10cm-1, 3121.25cm-1, 2996.13 cm-1, 1633.91 cm-1,were due to NH, NH2 , C=N and C=C functionalities, the 1H NMR spectrum showed peaks at δ 7.72 – δ 6.40 for aromatic protons while the peak at δ 4.72 was for the two proton of amine functionality. In the C NMR spectrum of compound 13 the peaks at 148.06 – 118.47 were assigned to the twelve aromatic carbons present in the compound. The molecular ion [M+] has a peak at 258.08. Finally for compounds 15a-c, the FTIR spectra (cm-1) showed peaks in the range 1385.04 to 1358.85 for NH stretch, 3121.78 to 3047.63 for CH aromatic stretch, 1626.05-1625.61 for C=N, C=C and 1385.04 -1358.85 for S=O vibrations. Their 1H NMR spectra (δ) had peaks in the range 9.00 to 8.36 for NH protons, 8.37 – 6.38 for aromatic protons and 2.73 for methyl protons while their 13C NMR spectra (δ) showed in the range 151.26 to 111.63. The compounds showed the following molecular ion peaks (m/z) at 399.44, 413.47 and 444.44 respectively in their HRM spectra.
3.2 In silico antimicrobial studies
The result for the antimicrobial in silico studies is presented in the table 1 below.
Table 1 Binding energies (kcal mol-1) for antimicrobial in silico studies
| Compounds | S. typhi 7D8I | S. aureus 7R1M | E. coli 7AB4 | B. cereus 4YXB | fumigatus 5ZVP |
| 13 | -7.2 | -6.4 | -6.0 | -7.2 | -6.6 |
| 15a | -8.0 | -7.5 | -7.0 | -7.4 | -5.9 |
| 15b | -8.3 | -7.5 | -7.1 | -7.9 | -6.2 |
| 15c | -8.8 | -7.5 | -7.6 | -8.6 | -6.9 |
| Ciprofloxacin | -8.3 | -6.4 | -6.2 | -8.1 | -6.9 |
| Ketoconazole | -8.0 | -6.4 | -8.0 | -7.0 | -7.6 |
The results on table 1 revealed compounds 15c with the highest binding energies of (-8.8, -7.5, -7.6, and-8.6) kcal mol-1 for the bacterial strains (S. typhi, S. aureus, E. coli and B. cereus) as well as -6.9 kcal mol-1 for the fungus(A.fumigatus)comparable to the standards, ciprofloxacin and ketoconazole respectively. From this observation, it could be seen that the compounds exhibited better interactions with the bacterial strains protein targets than their fungus counterpart implying better antibacterial properties than antifungal properties. Furthermore, compound 13 without the sulphonamide functionality exhibited a lower binding energies relative to the sulphonamide derivatives and the standard drugs. This observation corroborated with the claim that sulphonamide compounds has inhibition effect in the production of folic acid in bacteria cell wall responsible for their survival 15, 17. Figure 3 is a representative diagram displaying the interactions of hydrogen bonding of compound 15 c with active site of E. coli amino acid residues at ASPA: 73. Other interactions identified include Van der Waals, Water hydrogen bond, Pi-Anion, Pi-Akyl, carbon hydrogen bond.
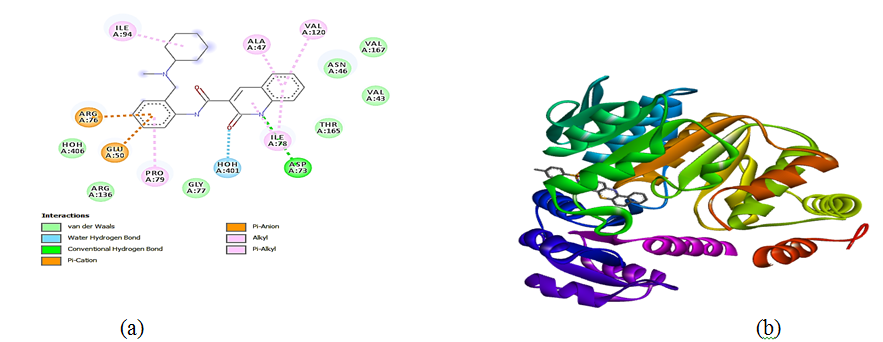
Figure 3: (a) 2D representation portraying hydrogen bonding and other interactions between 15c and the active site amino acids residues E. coli bound (b) 3D representation displaying interactions of compound 15c and the functional pockets of 7D8I protein target for S. typhi, which gave the highest binding energy of -8.8 kcal/mol
3.3 Drug likeness properties of synthesized compounds
The molecular parameters used to predict drug-likeness of synthesized compounds are presented in table 2.
Table 2: Drug likeness properties of the new derivatives
| Compounds | MW | mLogP | TPSA | HBA | HBD | Nviolations | Nrotb |
| 13 | 259.28 | 1.74 | 109.17 | 2 | 2 | 0 | 1 |
| 15a | 399.44 | 2.36 | 137.70 | 4 | 2 | 0 | 4 |
| 15b | 413.47 | 2.59 | 137.70 | 4 | 2 | 0 | 4 |
| 15c | 444.44 | 2.32 | 183.52 | 6 | 2 | 1 | 5 |
| Acceptable threshold | ˂500 Da | <5 | ≤140 A2 | ≤10 | ≤5 | 0 | 9 |
N.B. MW = Molecular Weight, TPSA = Topological Polar Surface Area, HBA = Hydrogen Bond Acceptor, HBD = Hydrogen Bond Donator, Nrotb = Number of rotable bonds, Nviolations = Number of violations, miLogP = Modified molecular hydrophobicity potential, 24.
According Lipinski et al, 24 ‘the rule of 5’ (RO5) predicts that for compound to qualify as a good drug molecule there should not be more than five hydrogen bond donors, ten hydrogen bond acceptors, the molecular weight must not be greater than 500 g/mol, the calculated Log P must not be greater than 5 24. As presented in Table 2, all the compounds obeyed the Lipinski rule of 5 (RO5) which suggest that a drug molecule must have a molecular weight that is less 500 Da (gmol-1), Log P value of less than 5, HBD and HBA values must be less than 10 and 5 respectively. However the rule was violated compound by 15c with regards to TPSA threshold of ≤140 A2 by compound 15c, which calls for structural optimization. TPSA value tells much about the permeation ability of a drug molecule in a cell membrane such as the GIT and the blood-brain barrier (BBB) 25. The lesser the TPSA value, the better its chances of being a good drug molecule 26, 27. Researches have shown that an increase in the value of TPSA reduces the mobility of a drug in the body and consequently diminishes its biological activities as well 28.
3.4 In vitro studies
The results for the in vitro studies showing the zones of inhibition of the new compounds against Salmonella typhi, S. aureus and A. fumigatus are presented in table 3 below.
Table 3. Zone of inhibitions of the derivatives against the microorganisms (mm)
| Compound | S. typhi | S. aureus | E. coli | A. fumigatus |
| 13 | 18.0 | 12.5 | 13.5 | 10.0 |
| 15a | 15.5 | 17.0 | 20.6 | 10.1 |
| 15b | 10.0 | 14.5 | 19.7 | 16.5 |
| 15c | 10.5 | 33.5 | 21.6 | 15.0 |
| Ciprofloxacin | 20.5 | 20.0 | 19.2 | NA |
| Ketoconazole | NA | NA | NA | 16.5 |
Note: 10.0-14.5 mm = low sensitivity, 15.0-20.5 mm moderate sensitivity and 21.0-33.5 high sensitivity, NA = Not applicable
The result of sensitivity test as presented in table 3, showed that the compounds exhibited varying degree of inhibitions for both the bacteria and the fungus strains relative to the standards. However, the bacteria showed more sensitivity than the fungus with 15c exhibiting the highest zone of inhibition against S. aureus (33.5mm) while compound 15b exhibited a zone inhibition of only 16.5 mm for the fungus, A. fumigatus (16.5mm) compared to the standard drug. This observation corroborated with the antimicrobial in silico studies presented in table 1 which portrayed compounds with sulphonamide functionalities as promising antibacterial agents because of their ability to interfere with folic acid synthesis, a key factor for bacterial survival when sulphonamide is mistaken by bacteria for p-aminobenzoic (PABA) 29. This is because the formation of dihydrofolic acid 20 which occurs when dihydropteridine diphosphate 18 reacts with glutamic acid 19 in (Scheme 2A) cannot take place when sulphonamide 5 is involved in (Scheme 2B)30. This observation clearly shows that sulponamide compounds exhibit bacteriostatic rather than bactericidal effect 31. These observations are illustrated in the scheme shown below
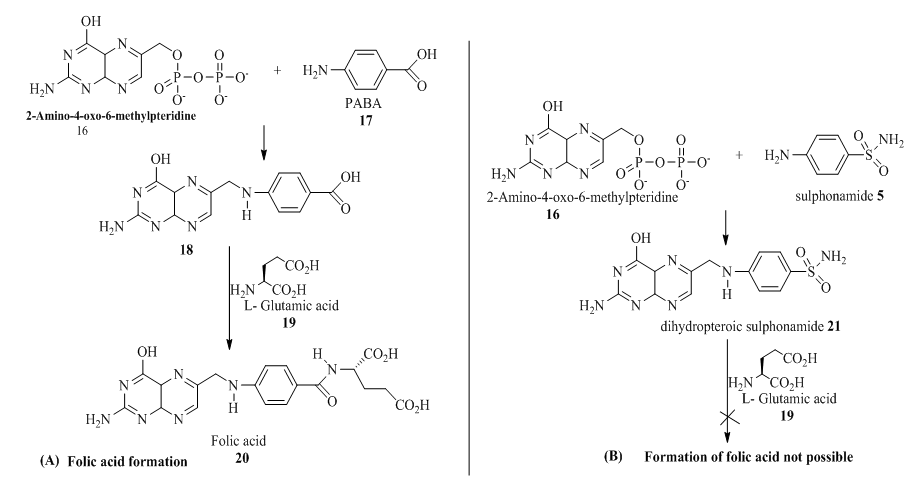
Scheme 2A & 2B: Synthesis of folic acid by PABA and its inhibition by sulphonamide
4.0 Conclusion
The synthesis of 7-nitro-10H-phenothiazin-3-sulphonamide derivatives were successfully carried out. The key intermediates, N1-(4-nitrophenyl)benzene-1,4-diamine11 and 7-nitro-10H-phenothia-zin-3-amine 12 were also successfully synthesized. 7-nitro-10H-phenothiazin-3-amine 13 was treated with three arylsulphonyl chlorides to furnish the three expected derivatives 15a, 15b, and 15c.The FTIR, 1HNMR, 13CNMR and HRMS results of the compounds agreed with their structures. The in silico antimicrobial studies showed compounds 15c with binding energies of (-8.8, -7.5, -7.6 and 8.6) kcal mol-1 for S, typhi, S. aureus, E. coli and B. cereus as well as -6.9 kcal mol-1 for the fungus(A.fumigatus)comparable to those of the standards, ciprofloxacin and ketoconazole respectively. Furthermore, compounds 15b and 15c exhibited the highest zones of inhibition (16.5 and 33.5) mm for the fungus, A. fumigatus and for the bacteria S. aureus comparable with the standard drugs. The results obtained revealed that these new compounds have promising biological properties. However, this studies was limited by a number of factors such as time and resources. We therefore recommend that further works such as cytotoxicity studies, in vivo evaluation, or pharmacokinetic profiling of the new derivatives be carried out.
References
[1]. Rani, J.V., Konda, R.K., Surekha, N.Y. Synthesis, Characterization and In-vitro Anti-Inflammatory Activity of Phenothiazine Derivatives. American Journal of Pharmtech. Rese. 2020; 10(1); 17-24.
[2]. Arun K, Chiara V, Donatella B, Marco L., Deepak K. ,Phenothiazines as anti-cancer agents: SAR overview and synthetic strategies, Eur J. Med. Chem. 254, 2023, 115337
[3]. Okafor C. O. Chemistry and applications of angular phenothiazines. Dyes and Pigments 1986; 7 (4), 249-287,
[4]. Venkatesan K, Satyanarayana V. S, and Sivakumar A. Synthesis and Biological Evaluation of Novel Phenothiazine Derivatives as Potential Antitumor Agents, Polycyclic Aromatic Comp. 2022, 1-11
[5]. Nourah A. Al Zahrani, Huda A. Al-Ghamdi, Reda M. El‐Shishtawy, Phenothiazine derivatives: Synthesis, docking studies and antimicrobial activity, J. Mol Struc, 1324, 2025, 140885,
[6]. Mayur M. Kishor H. Transition metal-free synthesis and functionalization of phenothiazines. Tetrahedron 145, 2023, 133618
[] B. E. Ezema, C. O. Okafor, C. G. Ezema and A. E. Onoabedje. Synthesis of new diaza angular and tetraazacomplex phenothiazine rings. Chem. Process. Eng. Res, 2012; 3, 40-47.
[8]. Faizal, K., Rajneesh, M. Recent advances in the development of phenothiazine and its fluorescent derivatives for optoelectronic applications. J. Mat. Chem. 2023; 11, 2786-2825.
[9]. U. C. Okoro and A. O. Ijeoma; Synthesis of new non-linear polycyclic diazaphenothiazine ring system. Int. J. Chem. 2006; 16, (4), 245-250.
[10]. Alexander I. K, Sofia O. S, and Yulia H. B, Electrochemical and photochemical functionalization of phenothiazines towards the synthesis of N-aryl phenothiazines: recent updates and prospects. Eur. J. Org. Chem. 2025, 1 – 6.
[11]. Dhanaraju, G.C., Magharla, D.D .Resent Progress in Synthesis, Structure and Biological Activities of Phenothiazine Derivatives. Rev. J. Chem. 2019; 9(2), 95-126.
[12]. Seeman, M. V. History of the dopamine hypothesis of antipsychotic action. World J. Psych. 2021; 11(7), 355.
[13]. Farah, Y., Oussama, M. and Jehad, H. Sulphonamides: Historical discovery development/structure-activity relationship notes. In-vitro In-vivo In-silico J. 2018; 1 (1):1-15
[14]. Ali, M., Angeli, A., Bozdag, M., Carta, F., Capasso, C., Farooq, U. and Supuran, C. T. Benzylaminoethylureido-tailed benzenesulphonamides show potent inhibitory activity against bacterial carbonic anhydrases. Chem. Eur. 2020; 15, 2444.
[15]. Khulood, H. O., Mazin, N. A. A., Ali, B. R., Hussein, A. A. and Fadi, A. M. The recent progress of sulfonamide in medicinal chemistry. Syst. Rev. Pharm. 2020; 11(12): 1473-1477.
[16]. Gajanan, S. and Vagdevi, H.M. Synthesis and invitro antibacterial activity of novel substituted N-1, 3-benzoxazol-2yl benzene sulphonamide. Int. J. Sci. Res. 2018; 7(12), 715-717.
[17]. Mussi, S., Rezzola, S., Chiodelli, P., Nocentini, A., Supuran, C. T., and Ronca, R. Antiproliferative effects of sulphonamide carbonic anhydrase inhibitors C18, SLC-0111 and acetazolamide on bladder, glioblastoma and pancreatic cancer cell lines. J. Enzy. Inhib. Med. Chem., 2022; 37(1), 280-286.
[18].Prakash, P. Combinatorial Synthesis: An Introduction. Acta Sci. Pharm. Sci. 2018; 2(4), 1.
[19]. Wang H-G, Williams R. E. & Lin P. F. A novel class of HIV-1 inhibitors that targets the viral envelope and inhibits CD4 receptor binding. Cur. Pharma. Des. 2004; 10 (15): 1785–1793
[20]. Song, L. and Ji, Q. The crystal structure of Staphylococcus aureus CntA in apo form. Protein Data Bank. Accession code 5YH5. (2018). Retrieved from https://doi.org/10.2210/pdb5YH5/pdb
[21]. Mima, M. and Ushiyama, F. Crystal structure of E.coli DNA gyrase B in complex with 2-oxo-1,2-dihydroquinoline derivative. Protein Data Bank. (2020). Accession code 6KZV. Retrieved from https://doi.org/10.2210/pdb6KZV/pdb
[22]. Trott, O. and Olson, A. J. (2010). AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading, Journal of Computational Chemistry, 31, 455-461. https://doi.org/10.1002/jcc.21334
[20]. Lipinski, C. A. Lead-and drug-like compounds: the rule-of-five revolution. Drug Discovery Today: Tech. 2024; 1(4), 337-341.
[23]. E. L. Ayuk, M. O. Uchegbu, P. I. Ebiem-Kenechukwu, and T. O. Oni. Synthesis, characterization, in silico and in vitro antimicrobial activity of phenothiazine-3-sulphonamide derivatives. Bioactivities. 2024; 2 (1), 41-56.
[24]. Lipinski, C. A., Lombardo, F., Dominy, B. W., & Feeney, P. J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Del. Rev. 1999; 23(1-3).
[25] T. O. Oni, E. L Ayuk. Synthesis of substituted arylsulphonamoyl ‘Leu-Gly’ dipeptide carboxamide derivatives containing antimalarial and other related diseases pharmacological properties. J. Mol. Struc. 1332 (2025) 141540
[26]. E. L. Ayuk, M. O. Uchegbu, P. I. Ebiem-Kenechukwu, T. O. Oni. Synthesis, characterization, in silico and in vitro antimicrobial activity of phenothiazine-3-sulphonamide derivatives. Bioactiv. 2 (1) (2024). 41-56. https://doi.org/10.47352/bioactivities.2963-654X.215
[27]. S. M. A. Kawsar, M. A. Hosen, Y. E. Bakri, S. Ahmad, S. T. Affi, S. G. Said. In silico approach for potential antimicrobial agents through antiviral, molecular docking, molecular dynamics, pharmacokinetic and bioactivity predictions of galactopyranoside derivatives, Arab J. Basic App Sc, 29, (1) (2022), 99-112, https://doi. org/10.1080/25765299.2022.2068275
[28]. S. M. A. Kawsar, A. Kumer, N. S. Munia, M. A. Hosen, U. Chakma, S. Akash. Chemical descriptors, PASS, molecular docking, molecular dynamics and ADMET predictions of glucopyranoside derivatives as inhibitors to bacteria and fungi growth. Org. Commun. 15 (2), (2022) 184-203, http://doi.org/10.25135/acg.oc.122.2203.2397
[29]. A. W. Hosny, M. A Gomha, S. M Zaki, E. A Magdi, B. E. Farag, F. A. Ahmed, A. Zaki, H. Yasser. Synthesis, molecular docking study, and biological evaluation and of new thiadiazole and thiazole derivatives incorporating isoindoline-1, 3-dione moiety as anticancer and antimicrobial agents. Res. Chem. 7(2024), 101375.
[30]. Philip A. Masters, Thomas A. O’Bryan, John Zurlo, Debra Q. Miller & Nirmal Joshi. Trimethoprim-sulphamethoxazole revisited, Arch Int. Med, 2003, 163 (1), 402-410
[31]. Hiren H Variya, Vikram Panchal & G R Patel. Synthesis, anti-tuberculosis and anti-bacterial activities of sulphonamide bearing 4-((2-(5-bromo-1H-pyrazolo-[3,4-b]pyridin-1-yl)-2-oxoethyl)amino)-N-(various substitutions)benzenesulphonamide, Indian J. Chem. 2020, 59 (1), 682-689
