Introduction
Bladder cancer (BC) is a disease of remarkable morbidity and mortality, characterized by high recurrence rate, pathological progression, and poor survival in advanced metastatic disease [1, 2]. BC accounts for nearly 30% of all cancers reported in Egypt, and this is accompanied with a high mortality rate relative to percentages reported in several countries in Europe and in the USA[3, 4]. Over 90 percent of the bladder tumors are urothelial carcinoma (UC), while squamous cell carcinoma (SCC) accounts for only five percent of BC in industrialized countries but represents more than 50percentof tumors presenting in Africa and the Middle East, where bilharzia (Schistosoma haematobium) is endemic [1].
Several factors are implicated in the cause of BC including cigarette smoke, occupational exposure to aromatic amines, arsenic in drinking water, urinary tract infections, medical conditions, and diet [2, 5-7].
Anticancer drugs are available for the clinical treatment of BC such as cisplatin and paclitaxel. They are administered via different techniques including in travesical, neoadjuvant, and adjuvant. The choice of a specific regimen depends on the type, location, and the stages of BC[8, 9]{Subramanian, 2015 #53;Abbasi, 2011 #118}.However, chemotherapy is always associated with severe side effects such as hair loss, suppression of bone marrow, drug resistance, gastrointestinal lesions, and cardiac toxicity, resulting from the untargeted nature of the drug [10, 11]. The other major drawback of chemotherapeutic regimes is the cost.With the high recurrence rate of BC which requires multiple treatments over prolonged periods of time, there is an urgent need to develop and/or discover alternative affordable, lower toxicity drugs [8, 12]. Nature has been the inspiring source for the discovery of new pharmaceuticals throughout several years[13]. For example, the Chinese used to drink the hot water extract of Artemisia for the treatment of malaria for thousands of years, and earlier this century the antimalarial artemisin was isolated and identified from Artemisia[14]. Green tea,whichis produced from the non-fermented leaves of the evergreen shrub Camellia sinensis plant, is another example of a healthy natural extract that the Chinese have been using for a long period of time to cure several symptoms including infections, high cholesterol levels, impaired immune function, cardiovascular diseases and recently cancer prevention [15, 16]. Green tea leaves are rich in bioactive compounds,in which polyphenols are the most active constituents[16]. The largest group of green tea polyphenols is a type of flavonoid, which account for more than 70 percent of total phenolic content. Flavanols(subclass or catechins) are the most plentiful compounds that account for 30-40 percent of dry weight of the leaves including epicatechin (EC), epigallocatechin (EGC), epicatechin gallate (ECG) and epigallocatechin Gallate (EGCG) (Figure 1) [15, 17, 18].
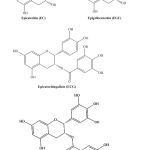 |
Figure 1: Chemical structure of the most abundant catechins in the leaves of the green tea. |
EGCG,in particular has been shown to possess numerous biological activities, such as anti-cancer activity [15]. Mechanic studies have indicated that EGCG interacts with cancer cells and hence exerts the cytotoxic effect against different targets. For example, EGCG was shown to suppress growth factor-mediated proliferation such as suppression of vascular endothelial growth factor (VEGF) and the epidermal growth factor receptor (EGFR) by modulating its protein tyrosine kinase activity[19].On the other hand EGCG was shown to inhibit several signaling pathways such as the nuclear factor-κB signaling pathway [20]. Evidences show that EGCG can be beneficial in treating brain, prostate, cervical, colorectal, and BC. It kills cancer cells without harming healthy tissue [21]. Although EGCG has been tested and studied in several cancer cell lines, no EGCG studies were found on an Egyptian-based BC cell lines. Based on our interest to develop and discover alternative treatment for BC at the Mansoura Urology Unit, Mansoura University, we report herein the anticancer evaluation and detailed molecular studies of EGCG on the Egyptian BC cell line (SCaBER).
Materials and Methods
The human bladder squamous carcinoma cell line (SCaBER cell line) was obtained from Tumor Biology Department, National Cancer Institute of Cairo University. SCaBER cells were cultured in Dulbecco’s modified eagle’s medium pH 7.2, low glucose (DMEM) that was purchased from Sigma-Chemical Co. (St. Louis, MO, USA) supplemented with 20% (v/v) fetal bovine serum (FBS), 100 (IU/ml) penicillin, and 100 (µg /ml) streptomycin (Invitrogen Life Technologies). Cells were cultured at a density of 4×10^4 / cm2 in a humidified atmosphere of 95% air and 5% CO2 incubator at 37°C. Once the cells reached approximately 80% confluency, they were sub-cultured using trypsin (0.25%), then expanded onto more culture flasks.
Experimental Treatments
EGCG (purity >95%), obtained from Sigma Aldrich (St. Louis, MO, USA), was dissolved in distilled water to make a stock concentration of 11mMfollowed by filtration using sterile syringe filter 0.2μm (corning incorporator, Germany), and then, stored in aliquots at -20 °C. To evaluate the anticancer and dose dependent effects of EGCG on cell growth, cells were seeded in triplicates in 6-well plates at a density of 5×105 cells per dish and grown for 24 hours. Cells were then incubated with 0.011 µM, 0.027 µM, 0.055 µM, 0.11 µM and 0.22 µM of EGCG while control cultures contained DMEM medium for 24, 48, and 72 hours.
Morphological Study
Cultured cells at each concentration andtime point of EGCG were imaged three times with OLYMPUS inverted microscope model LX71S1F-3.(Tokyo, Japan.)
Viability Test
Cell viability is the percentage of viable cells among the total (dead and viable). To measure viability of the cells, triplicate wells of cells for each concentration were counted after The trypsinized cells were harvested with the suspended cells and transferred into a 15 ml Falcon tube, and then the cells were stained with an equal volume of 4% trypan blue. The number of dead and viable cells was counted three times by hemocytometer.
Molecular Biology Study
Isolation of Total RNA
Total RNA was isolated from each sample of 5 ×105SCaBER cells which were completed according to the manual instructions of RNeasy Plus mini kit (QIAGEN Corporation Hilden, Germany). Then, the concentration of the isolated RNA was assessed using Nano Drop 2000 (Thermo scientific, USA), at an OD of 260 nm. RNA samples with OD A260/A280 ratios within 1.9-2.1, and A260/230 ratio within 1.8-2 were used for qRT-PCR. To ensure RNA integrity, denaturing agarose gel electrophoresis (1.5%) was performed to detect two distinct characteristic RNA bands. After that, 1.5 µg of RNA was reverse-transcribed to cDNA by RT2 First Strand kit (Qiagen Sciences, Maryland, USA).
Table 1: Primer sequences used in RT-PCR analyses
|
Gene |
Forward primer |
Reverse primer |
Annealing temperature
(Cº) |
PCR Products
(bp) |
| UPII | AGCAAAGTGGTGACGTCCAG | TGTGAAGCCAGCACCACTGT | 58 | 90 |
| EGFR | TCTGGAAGTACGCAGACGCC | GGACGGGATCTTAGGCCCAT | 60 | 116 |
| Surviving | CCCCACTGAGAACGAGCCAG | TGTTCCTCTATGGGGTCGTCA | 60 | 93 |
| GAPDH | CGCTGAGTACGTCGTGGAGT | GGGGGCAGAGATGATGACCC | 60 | 100 |
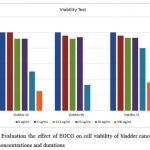
Table 2: The percentage of viable cells at different concentrations (μM) and durations in hours.
| duration | Concentration% | |||||
| 0µM | 5µM | 12.5µM | 25µM | 50µM | 100µM | |
| 24 | 100 | 100 | 92 | 92 | 50 | 36 |
| 48 | 100 | 96 | 92 | 94 | 33 | 25 |
| 72 | 100 | 96 | 93 | 93 | 80 | 0 |
Quantitative Real-Time PCR (qRT-PCR)
Relative gene expression was measured by Real Time PCR Instrument (CFX96, BIO RAD, USA).Each sample was performed in triplicate. The reaction was done by 12.5 μLof 2X SYBR Green PCR Master Mix (Qiagen),2μL of cDNA template, 2 μL(10pmol) gene primer and 10.5 μL of RNase-DNase free water. The primer sequences were designed online using the NCBI site and manufactured at Vivantis (Selangor, Malaysia) as shown in table 1.
Reaction mixture is subjected to Real-Time PCR machine using the following conditions: The first stage required 50◦C for 2 minutes. The second stage needed 94◦C for 5 minutes. The third stage had 40 cycles of amplifications that were performed by the denaturation step at 94◦C for 30 seconds, annealing step temperature according to each primer for 30 seconds and an extension step at 60◦ C for 30 seconds. Melt curve analysis was used to determine the specificity of the PCR products. Samples were normalized to glyceraldehyde 3-phosphate dehydrogenase GAPDH. Samples were calculated relative to that of negative controls (0µM)and according to the mathematical model of R = 2- ΔΔCt [22].
Results
The induction of apoptosis by EGCG was confirmed morphologically by examination of SCaBER cells treated with EGCG under inverted microscope. The changes have been observed are cell shrinkage, nuclear condensation, and formation of pyknotic in contrast the untreated cells Figure (2).
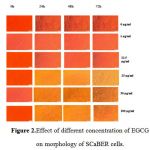 |
Figure 2: Effect of different concentration of EGCG
|
Viability Test
The effect of EGCG at a dose of 0.011 µM, 0.027 µM, 0.055 µM, 0.11 µM and 0.22 µM on viability of the cell line with different treatment duration (24, 48, and 72hrs) is shown in Table 2. A remarkable viability loss was observed of SCaBER cell line after treatment with EGCG especially with high concentrations, (P < 0.0001), as shown in Figure (3).
Gene Expression by Real Time PCR
Relative qRT- PCR was performed for UPII, EGFR and surviving genes to determine the fold difference of a certain mRNA level between different samples.All tested genes were abundant and showed single peaks in melting curve analysis with a single band in high-resolution of 2% agarose gel electrophoresis. Gene expression was normalized to the house keeping gene GAPDH, whose expression remained constant during the entire study and was not affected by EGCG concentrations. We found that EGCG suppresses the expression of UPII, EGFR and surviving in a time and dose-dependent manner as shown in figures(4, 5, and 6).
 |
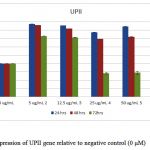 |
Figure 4: Expression of UPII gene relative to negative control (0 μM) |
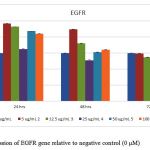 |
Figure 5: Expression of EGFR gene relative to negative control (0 μM) |
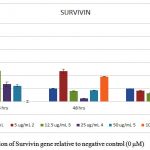 |
Figure 6: Expression of Survivin gene relative to negative control (0 μM) |
Discussion
The anticarcinogenic properties of green tea’s extract and its individual natural products, particularly EGCG, have been extensively studied in different types of cancers both in vitro and in vivo, such as gastric cancer [23], ovarian cancer [24], cervical cancer [25], hepatocellular cancer [26], and BC [27-29]. Several studies suggest that green tea ingredients are a good approach for the prevention and treatment of BC [29]. Suganuma, Okabe [30] found that greentea’scatechins were widely distributed through the mouse body and concentrated in the bladder after green tea ingestion.
At the current study, microscopic examination of BC cell line (SCaBER) treated with different concentration of EGCG revealed some morphological changes including cell shrinkage, nuclear condensation, and formation of pyknotic bodies of condensed chromatin, relative to untreated cells.Such changes increased significantly with increasing EGCG concentration and duration of its exposure to cells. Those changes are reported previously as characteristic features for apoptosis [29,31]. That observation goes hand-in-hand with the publication of Rao and Pagidas [24] on the human ovarian cancer cell line, as well as with Sharma, Nusri [25] experiments on human cervical cancer cell line treated with EGCG.
A significant reduction of the cell’s viability (P ≤ 0.001) was observed with respect to EGCG concentrations and duration of cell exposure which clearly suggests that EGCG is inhibiting the growth of the bladder squamous cell line at appropriate concentration.Lee, Nam [31] and Liu, Hou [32] data on squamous cell carcinoma on head, neck and esophagus also confirmed the inhibitory effect of different EGCG concentration on cell line growth and proliferation. However, findings of Mayr, Wagner [33] and Qin, Wang [27] on biliary cell carcinoma and bladder cell carcinoma respectively emphasize that the effect of EGCG on cell line is concentration dependent.
Chen, Wan (36)has shown that EGCG was able to target several biological pathways such as gene expression.Thus, we explored the effect of EGCG on molecular targets that can be used for cancer screening, early diagnosis, in addition to prediction of disease recurrence, and survival such as UPII, EGFR and surviving[36-38].
Uroplakins are cell membrane proteins synthesized as a principal product of human urothelial cells differentiation and turned out to be cancer cell markers [39,40]. A significant down-regulation of UPII mRNA (P ≤ 0.001) associated with high concentrations of EGCG especially after 72 h incubation, has been observed indicating that there was growth inhibition of cancer cells.
Further more,over expression of EGFR in BC has been linked to essential carcinogenesis process such as uncontrolled cell proliferation, cell cycle progression, angiogenesis increase and apoptosis reduction [41-43]. In the current study, EGFR expression was reduced significantly (P ≤ 0.001) in a dose and time dependent manner, which revealed that EGCG may be an inhibitor for EGFR. Therefore EGCG may have effective potential in therapeutic value. These results can relate with the results of Ma, Li [44] on lung cancer cell in addition to results of Chang, Chang [45] on human oral squamous carcinoma cells. EGCG was suggested to inhibit the EGFR signaling pathway through the interruption of EGFR’s ability to bind its receptor [46,47].
The last studied marker was surviving which is regarded as a structure unique inhibitor for apoptotic family, and is encoded by BIRC5. Over expression of surviving has been associated with apoptosis inhibition, promotion of proliferation, and angiogenesis. Surviving is highly expressed in several malignant tissues, such as bladder, when compared to normal tissues [38,48,49]. Thus, great concern has been attracted to surviving as a principle target for antitumor therapy and as a potential biomarker for cancer [50,51]. In this study, the gene expression of surviving undergoes a significant down regulation(P ≤ 0.005) by increasing EGCG concentration. These results align with the results of Tang, Zhao [52] in the effect of green tea extract on breast cancer cells who stated that this down regulation is a key step in the molecular mechanism involved in the anti-cell proliferative and effect of green tea extract. Moving on, this finding is supported with the study conducted by Onoda, Kuribayashi [53] who confirmed that EGCG induces apoptosis in gastric cancer cell line by down regulation of surviving expression and hypothesized that surviving may be a molecular target of EGCG.
In conclusion EGCG, a green tea constituent, has the ability to inhibit the growth of cancer cells in vitro through proliferation discouragement and induction of apoptosis. Subsequently, EGCG could be a potentially cheap and available anticancer agent in bladder squamous cell carcinoma
Acknowledgments
We wish to thank Dr. Sawsan El Halawani and Dr. Romaila Raouf for kindly providing the necessary facilities to complete this study and for their encouragement and support. Nandita Kumar for revising the manuscript. This work was supported by Urology Nephrology Center Mansoura University
Conflict of interest
The authors declare that there is no conflict of interests regarding the publication of this paper.
References
- Yazbek-Hanna M, Whelan P, Jain S, Bladder Cancer Medicine, 2016.
- Burger M, Catto JW, Dalbagni G, Grossman HB, Herr H, Karakiewicz P, Eur.Urology;63(2),234. 2013.
- Barh D, Carpi A, Verma M, Gunduz M. Diagn. Progn:, CRC Press; 2014.
- Rambau P F, Chalya PL, Jackson K., Infectious. Agents and Cancer. 8(1),19;2013
- Lunt C, Maddineni S, Brough R, British J. Med. Surg. Urology. 5(2),95.2012
- Amr S, Dawson R, Saleh DaA, Magder LS, Mikhail NN, St. George DM, Arch. Environ.Occup.Health. 69(1),3;2014
- Jankovic S, Radosavljevic V, Tumor. 93(1),4; 2007.
- Subramanian A, John A, Vellayappan M, Balaji A, RSC Advances. 5(45):35608-21;2007
- Kaufman DS, Shipley WU, Feldman AS, Bladder Cancer. The Lancet. 374(9685),239. 2009.
- Ghorbani A, Hosseini A, Avicenna J. Phytomed. 5(2),84;2015.
- Shanmugam MK, Lee JH, Chai EZP, Kanchi MM, Kar S, Seminars in Cancer Biology; Elsevier. 2016.
- Aliya S, Devi YP, Uma A, Current Trends in Biotech. & Pharm. 10(1);2016.
- Nobili S, Lippi D, Witort E, Donnini M, Bausi L, Mini E, Pharm.Res. 59(6),365;2009.
- Guo Z, Acta Pharm.Sinica B. 6(2);115;2016.
- Granja A, Pinheiro M, Reis S, Nutrients. 8(5),307;2016.
- Jigisha A, Nishant R, Navin K, Pankaj G, Intern .Res .J .Pharm. 3(5),139;2012.
- Lorenzo JM, Munekata PES, Asian Pacific J. Tropical Biomed. 6(8),709;2016.
- Tsao R, Nutrients. 2(12),1231;2010.
- Min K-j, Kwon TK, Integrative Med. Res. 3(1),16;2014.
- Shankar S, Ganapathy S, Srivastava RK, Front Biosci. 12(4881),51;2007.
- Makiuchi T, Sobue T, Kitamura T, Ishihara J, Sawada N, Iwasaki M, Cancer Sci. 107(1),76;2016.
- Pfaffl MW, Nucleic Acids Res. 29(9), 45;2001.
- Yang C, Du W, Yang D, Intern.J.Food Sci. and Nutr. 67(7),818;2016.
- Rao SD, Pagidas K, Anticancer Res. ;30(7),2519;2010.
- Sharma C, Nusri QE-A, Begum S, Javed E, Rizvi TA, Hussain A, Asian Pacific J.Cancer Prevention. 13(9),4815;2012
- Darvesh AS, Bishayee A,Nutr. and Cancer. 65(3),329;2013.
- Luo K-W, Chen W, Lung W-Y, Wei X-Y, Cheng B-H, Cai Z-M, Nutr. Biochem. 41:56-64; 2017.
- Qin J, Wang Y, Bai Y, Yang K, Mao Q, Lin Y, Molecular Medicine Reports. 6(5),1040;2012.
- R Conde V, G Alves M, F Oliveira P, M Silva B, Anti-Cancer Agents in Med. Chem. Formerly Current Med. Chem.-Anti-Cancer Agents. 15(1),26;2015.
- Suganuma M, Okabe S, Sueoka N, Sueoka E, Matsuyama S, Imai K, Molecular Mechanisms of Mutagenesis. 428(1),3394;1999.
- Sayed A SN, Abdul Wahab N, Abd Malek SN. In Vitro , Complementary and Alternative Medicine. 14;2013.
- Lee SH, Nam HJ, Kang HJ, Kwon HW, Lim YC, Eur. J. Cancer. 49(15),3210;2013.
- Liu L, Hou L, Gu S, Zuo X, Meng D, Luo M, Oncology Reports. 33(1),297;2015.
- Mayr C, Wagner A, Neureiter D, Pichler M, Jakab M, Illig R, BMC Complementary and Alternative Medicine.. 15(1),194;2015.
- Chen D, Wan SB, Yang H, Yuan J, Chan TH, Advances In Clin.Chem. 53:155;2011.
- Gabril MY, Yousef GM, Molecular Testing in Cancer: Springer. 301;2014.
- Gakis G, Schwentner C, Todenhöfer T, Stenzl A, BJU Intern. 110(2),233;2012.
- Jeon C, Kim M, Kwak C, Kim HH, Ku JH, A systematic Review and Meta-Analysis. PLoS One. 8(10),76719;2013.
- El Shobaky A, Abbas M, Raouf R, Zakaria MM, Ali-El-Dein B, Egypt. J. Basic and Applied Sci. 2(3),176;2015.
- Olsburgh J, Harnden P, Weeks R, Smith B, Joyce A, Hall G, J. Pathology. 199(1),41;2003.
- Badawy AA, El-Hindawi A, Hammam O, Moussa M, Helal NS, Kamel A, Current Urology. 9(4):192-201;2015.
- Mason RA, Morlock EV, Karagas MR, Kelsey KT, Marsit CJ, Schned AR, Carcinogenesis. 30(7),1155;2009.
- Izumi K, Zheng Y, Li Y, Zaengle J, Miyamoto H, Intern. J. Oncology. 41(5),1587;2012.
- Ma Y-C, Li C, Gao F, Xu Y, Jiang Z-B, Liu J-X, Oncology Reports. 31(3),1343;2014.
- Chang C-M, Chang P-Y, Tu M-G, Lu C-C, Kuo S-C, Amagaya S, Oncology Reports. 28(5),1799;2012.
- Kadioglu O, Cao J, Saeed M E, Greten HJ, Efferth T, Targeted Oncology. 10(3),337;2015.
- Yang CS, Wang H, Molecular Nutrition & Food Res. 55(6),819;2011.
- Garg H, Suri P, Gupta JC, Talwar G, Dubey S, Cancer Cell Intern.16(1),49;2016.
- Yang R, Liu M, Liang H, Guo S, Guo X, Yuan M, Molecular Cancer. 15(1),82;2016.
- Guindalini RSC, Machado MCM, Garicochea B., Molecular Diagnosis &Therapy. 17(6),331;2013.
- Hagen RM, Chedea VS, Mintoff CP, Bowler E, Morse HR, Ladomery MR, Intern. J. Oncology. 43(1),194;2013.
- Tang Y, Zhao DY, Elliott S, Zhao W, Curiel TJ, Beckman BS, Intern. J. Oncology. 31(4):705-11;2007.
- Onoda C, Kuribayashi K, Nirasawa S, Tsuji N, Tanaka M, Kobayashi D, Intern. J. Oncology. 38(5),1403;2011.
