INTRODUCTION
The genus Cymbidium Swartz is a member of the Orchidaceae family, which includes about 48–55 species and is primarily available in subtropical and tropical Asia1.Cymbidium orchids are impressive in worldwide horticulture because of their fragrant flowers and variegated leaves, and they have been grown for over ten centuries2-3. In Vietnam, there are 24 species, including 13 that grow on trees, 5 that grow in soil or rock crevices, 5 that cling to rocks, and 1 species that lacks leaves and survives in humus thanks to a highly developed root system. Vietnam, with its tropical and subtropical climate and numerous high-altitude mountainous areas, provides ideal conditions for the growth of many Cymbidium species. These orchids are widely distributed across the country, particularly in northern provinces such as Hanoi, Hung Yen, Ninh Binh, Sapa (Lao Cai), Vinh Phuc, Hoa Binh, Yen Tu, Lang Son, Cao Bang, and the Hoang Lien Son Mountain range. In the south, they are mainly found in the Central Highlands, especially in Da Lat (Lam Dong) and along the Truong Son Mountain range4.
Since the concept of DNA barcoding emerged and its applications, many countries worldwide focused on building DNA databases for native organisms. Orchids quickly became a subject of great interest among scientists due to their unique biological characteristics and economic and medicinal value5-6. The Orchidaceae family is considered one of the most challenging plant families to identify and classify, especially during the non-flowering stage. This presents significant difficulties in species identification, conservation, and evolutionary studies. The application of DNA barcoding has helped address these challenges, providing a more accurate method of species recognition compared to traditional morphology-based approaches7. Many species differ from their closely related counterparts by only very subtle and minor morphological characteristics, making it difficult to identify using conventional morphology-based methods. Furthermore, because Orchidaceae species can easily hybridize both in the wild and under cultivation, numerous intermediate forms and variations arise, making their classification at the genus and species levels extremely challenging8-10.
DNA barcoding has been widely used for numerous taxa of Orchidaceae in some previous studiessuch as Dendrobium11-15, Phalaenopsis16, Cypripedium3, Grammatophyllum17, Cymbidium18, Vanda19-20, Spathoglottis21, and Holcoglossum22. With the advent of DNA barcoding technology, it quickly became a powerful tool for identification of species, particularly in orchids8,23. DNA barcoding has been successfully applied in the classification and identification of orchid species. The psbA-trnH intergenic spacer in the chloroplast genome, along with the matK and rbcL gene sequences, have been successfully used to classify Dendrobium species 24-25. Additionally, matK DNA barcoding has effectively distinguished medicinal orchid species, including D. pulchellum, D. fimbriatum; D. nobile, D. moniliforme; and D. tosaense25. In the Cymbidium, four DNA regions for rpoC1, rpoB, trnH-psbA, and matK—were analyzed to identify species of Cymbidium in Thailand18. The sequences of these standard regions as barcodes were used for studies on genetic distances. The genetic distances mean nucleotide variations in tag sequences based on trnH-psbA, rpoB, matK, and rpoC1 and spacer region sequences of the 19 species ranging from 0.026 to 0.528, 0.012 to 0.546, 0.052 to 0.385, and 0.018 to 0.546, respectively. The phylogenetic relationships of Cymbidium were preliminarily applied using various molecular markers, including RAPD26, AFLP27, ISSR28-29, and SSR30. The matK and ITS regions were also used for examining the phylogenetic relationships among some polularl Cymbidium species31,29.
In this study, we aimed to test universality of a set of DNA regions in Cymbidium for identifying the species of Cymbidium and reconstructing the phylogenetic relationships within the genus Cymbidium. The matK gene was used to identify Cymbidium species and analyze the phylogenetic relationships among 16 Vietnam native Cymbidium species.
MATERIALS AND METHODS
Plant materials
Sixteen samples of Vietnamese native Cymbidium sp orchids (aged about 2-4 years)
species were collected in Quang Ninh, Lao Cai, Hoa Binh, Ha Noi, Lao Cai, and Da Lat provinces, and were grown at the nethouse of Institute of Agricultural Genetics, Hanoi, Vietnam.
DNA extraction
Genomic DNA was extracted from fresh leaves or flowers using the DNeasy Plant Mini Kit (QIAGEN, Hamburg, Germany), according to the manufacturer’s guidelines, and used as a template for PCR amplification. Genomic DNA was isolated from fresh leaf tissue with the DNA Plant Mini Kit (Qiagen). Briefly, the PCR reaction mixture (15µl) contained 1.5µl of 10x PCR buffer (with MgCl2), 0.2µl of dNTPs (10mM/µl), 0.1µl of Taq polymerase (5U/µl), and 1.5µl of forward and reverse primers (10pmol/µl). The primers used to amplify the matK region included matK2.1F forward primer (5’-CCTATCCATCTGGAAATCTTAG-3’) and matK5R reverse primer (5’-TCTAGCACAAGAAAGTCG-3’). A template of approximately 50-100 ng of genomic DNA was used for the reaction. The mixture was denatured at 94°C for 3 minutes, followed by 30 cycles at 94°C for 15 seconds, 58°C for 30 seconds, and 72°C for 30 seconds, with a final extension at 72°C for 7 minutes. The PCR products were analyzed by 1% agarose gel electrophoresis and visualized using ethidium bromide staining under UV light. DNA purification was carried out using the DNA Purification System (Qiagen Kit)32.
Sequencing peformance
The purified PCR products were directly sequenced by ABI PRISMTM 310 Genetic Analyzer (Applied Biosystems). The primers matK2.1F and matK5R for matK region were used for the sequence reaction. The matK region of each individual sample was sequenced in both the 5’direction and 3’direction at least twice to avoid mutation introduced by Taq polymerase. The boundaries of the matK region were calculated by comparing them with the sequences of the Orchids family species from the GeneBank.
Statistical analysis
The sequence analyses were aligned and compared using the ClustalW programs and analyzed using the MEGA 5.2.1 programs.
RESULTS AND DISCUSSION
Molecular makers to identify Vietnamese native Cymbidium based on matK region sequences
By using 2.1F và 5R primer pairs, we successfully amplified matK region by PCR products. The results showed that the PCR products were high quality with only one band in size ranging from 800-900 bp (Fig. 1). The band size was obvious, and correct size as expected, which should be able to be used for sequencing.
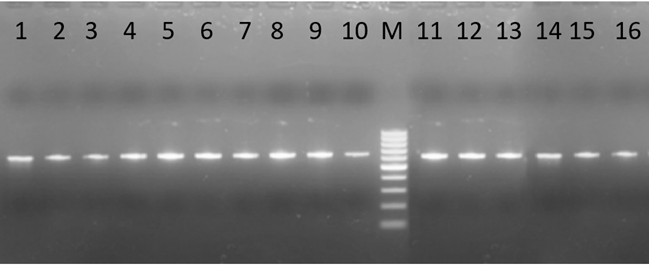
Fig 1. Electrophoresis of amplification matK segment on 16 Vietnamese native Cymbidium species samples by PCR. Lanes: 1, 4, 5, 6, 7, 8, 9, 10, 11, 12, 12, 14, 15, 16: 16 Vietnamese native Cymbidium samples;M: 100bp ladder
Analysis of sixteen Vietnamese native Cymbidium species based on matK sequences
Nucleotide Composition and DNA Sequence Length Vietnamese Native Cymbidium species
The results of sequencing showed that the sixteen Vietnamese native Cymbidium specieswere amplified and sequenced the matK total length of nucleotides obtained from 485 to 936 nucleotides with an average of 774.1 nucleotides as shown in Table 1.
Table 1. Nucleotide Composition and DNA Sequence Length Vietnamese Native Cymbidium species
| Names of samples | T(U) | C | A | G | Length of sequencing (bp) |
| Thanh Truong | 39.3 | 16.4 | 29.9 | 14.5 | 512.0 |
| Kiem lo hoi | 37.8 | 17.2 | 30.2 | 14.8 | 913.0 |
| Mac rung yen tu | 37.9 | 17.4 | 29.5 | 15.2 | 914.0 |
| Bach Ngoc Tieu Kieu | 39.6 | 16.6 | 29.8 | 14.1 | 604.0 |
| Bach Ngoc Dai Kieu | 38.9 | 16.8 | 30.3 | 13.9 | 719.0 |
| Bach Ngoc Xuan | 38.7 | 16.2 | 30.4 | 14.6 | 917.0 |
| Hong Hoang Sapa | 38.4 | 16.5 | 29.9 | 15.2 | 916.0 |
| Dai Mac la dung | 39.0 | 16.5 | 30.3 | 14.2 | 485.0 |
| Hoang Diem Vang | 38.9 | 16.6 | 29.5 | 15.0 | 519.0 |
| Dai Mac la cong | 33.0 | 25.6 | 22.8 | 18.6 | 848.0 |
| Thanh Ngoc | 38.5 | 15.4 | 29.5 | 16.7 | 936.0 |
| Dao Co | 37.6 | 14.9 | 30.0 | 17.4 | 919.0 |
| Mac Bien | 39.5 | 16.5 | 29.5 | 14.4 | 478.0 |
| Cam To | 37.5 | 16.9 | 29.8 | 15.7 | 915.0 |
| Tu Thoi | 38.6 | 16.7 | 29.4 | 15.2 | 914.0 |
| Tran Mong | 39.2 | 15.8 | 30.4 | 14.5 | 877.0 |
| Total | 38.3 | 17.0 | 29.5 | 15.3 | 774.1 |
The percentage of nucleotide as T (U) =38.3%; C = 17%; A =29,5%; and G = 15.3%. The nucleotide composition analysis reveals that thymine (T) and adenine (A) constitute approximately 67.8% of the total bases, while guanine (G) and cytosine (C) make up about 32.3%. This indicates a high AT content in the sequences, which is characteristic of the matK gene, known for its higher A-T content compared to G-C, facilitating genetic identification within the Orchidaceae family (TLTK). Comparing sequence lengths among the samples, the longest sequences are found in Bach Ngoc Xuan (917 bp), Hong Hoang Sapa (916 bp), and Dao Co (919 bp). The shortest sequences are observed in Dai Mac la dung (485 bp) and Mac Bien species (478 bp), with an average sequence length of approximately 774 bp across the samples. Samples with longer sequences (~914-919 bp) are likely species or varieties that are genetically stable and less affected by mutations. In contrast, samples with shorter sequences (~478-485 bp) may have undergone genetic variations or gene segmentations due to environmental factors or evolutionary differences.
Alignment of Vietnamese native Cymbidium species and Cymbidium species
in the world
To compare the difference between Vietnamese native Cymbidium species and
Cymbidium species in the world, we have conducted the analysis of the samples.
of Vietnamese species and the world based on the analysis coming aligned
columns. In this study, sixteen Vietnamese native Cymbidium species were alighted with 24 cymbidiums in GenBank. By the analysis of aligns upcoming column (alignments), The “Dai mac la cong” sample exhibits many unique SNPs that have not appeared in the other orchid samples, indicating that it has accumulated numerous distinct mutations during its evolution. This suggests that this sample may have diverged from other orchid lines very early, or it may carry characteristic mutations that set it apart evolutionarily from the others. Therefore, “Dai mac la cong” represents a valuable genetic resource, important for evolutionary and conservation studies, and it holds potential for use in breeding programs to enhance desirable traits in orchids (Fig 2). Our results were in line with the previous findings to accurately identify the genus of rosa species32.
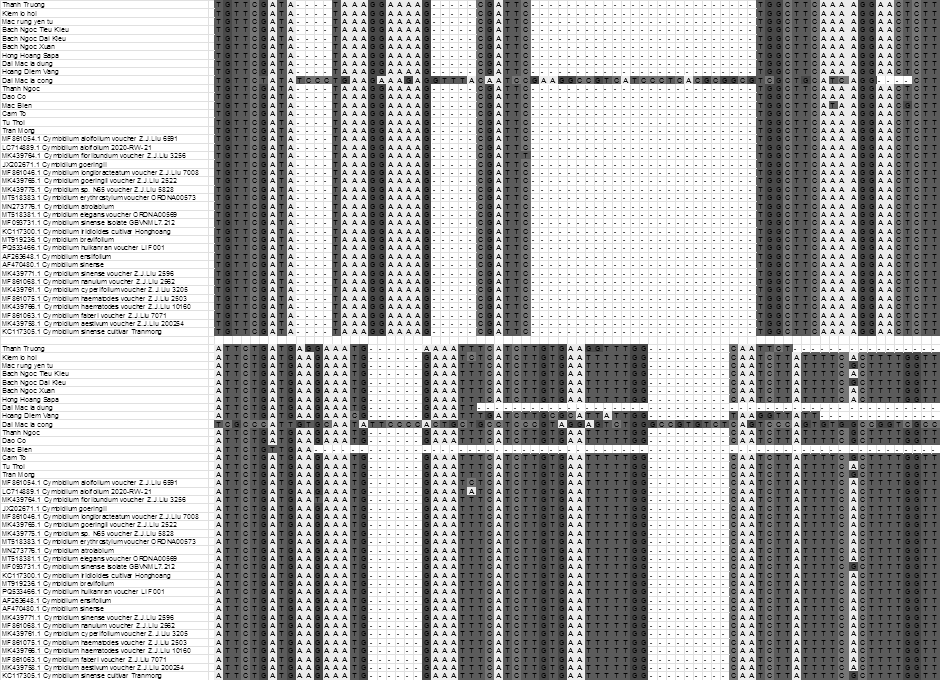
Fig 2. Alignment of Vietnamese native Cymbidium species and Cymbidium species in the world
Phylogenetic trees based on matK region sequences
Before launching the ITS database, it is important to construct the 16 species of Vietnamese Cymbidium species in this study, we collected 24 references matK sequences of Cymbidium species which are closely related to 16 species (Vietnamese Cymbidium) The alignment process was performed using the MEGA 5.2.1 programs software. The final dataset includes 40 sequences consisting of 24 control groups (cymbidium) with an analytical sample and 16 matK sequences from the analytical samples.
According to the phylogenetic trees, 16 Vietnamese native Cymbidium species trees were divided into 3 different groups (Fig 3).
Group I included 12 Vietnamese native Cymbidium and 21 Cymbidium species in the world and divided into 3 subgroups:
– Subgroup I.1: This subgroup includes 11 samples, consisting of five Vietnamese Cymbidium samples and six international Cymbidium varieties. The samples in this subgroup were included: Mac rung Yen tu, Dao Co, Bach Ngoc Dai kieu, JX202671.1 (Cymbidium goeringii) KC117305.1 (Cymbidium sinense cultivar), MT518383.1 (Cymbidium erythrostylum voucher ORDNA00573, )MK439758.1 (Cymbidium aestivum voucher Z.J.Liu 200254), AF470480.1 (Cymbidium sinense), Tran Mong, Dai Mac La Dung, Hoang Diem Vang, MF093731.1 (Cymbidium sinense), MF861046.1 (Cymbidium longibracteatum voucher Z.J.Liu 7008), MK439765.1 (Cymbidium goeringii voucher Z.J.Liu 2522), MK439775.1 (Cymbidium sp. N65 voucher Z.J.Liu 5828)
This group consists of closely related Cymbidium orchids, including common species such as Cymbidium sinense and Cymbidium goeringii, along with some local varieties. This branch may represent an ancient lineage of Cymbidium orchids, with some samples exhibiting significant evolutionary divergence. Mac rung Yen tu, Dao Co, Bach Ngoc Dai Kieu may form a localized group with close evolutionary relationships. These samples are positioned very closely together in the phylogenetic tree, indicating a strong genetic connection. This could be due to a shared origin or natural hybridization within a geographically close area. They may represent different variants of the same species, with minor morphological or ecological differences. These three samples are closely related to JX202671.1 (Cymbidium goeringii), suggesting a connection between the local samples and the widespread species Cymbidium goeringii.
Three samples such as Tran Mong, Dai Mac la dung, and Hoang Diem Vang are clustered within the same major branch as Cymbidium sinense and Cymbidium goeringii, indicating a close evolutionary relationship. These samples might belong to local lineages or hybrid varieties with some genetic differentiation. The bootstrap value of this branch is 5, indicating that the genetic differences among the samples are not very significant, possibly originating from a common recent ancestor. Trần Mộng is closely related to Cymbidium sinense within the group, potentially representing a variant or hybrid between C. sinense and another species in the same genus. It may have unique floral traits, although specific mutation characteristics are not well documented. Dai Mac La Dung is positioned near Tran Mong, suggesting a similar origin. It could be a form of Cymbidium sinense or a hybrid with Cymbidium goeringii. Hoang Điem Vang is in the same group as Dai Mac La Dung and Tran Mong but may have distinct genetic characteristics, possibly belonging to a local lineage of Cymbidium sinense with slightly different evolutionary traits. In summary, Tran Mong, Dai Mac La Dung, and Hoang Diem Vang have a close evolutionary relationship and may be variants or hybrids of Cymbidium sinense.
The Thanh Ngoc sample is evolutionarily related to species in the Cymbidium genus, particularly those connected to Cymbidium sinense and Cymbidium atropalabium. Thanh Ngọc has a moderate level of genetic differentiation compared to other samples and is grouped with MN273776.1 (Cymbidium atropalabium), AF263648.1 (Cymbidium ensifolium) and MT919236.1 (Cymbidium brevifolium). This group has a bootstrap value of 5, indicating a moderate level of phylogenetic support, suggesting that some differentiation still exists within the group. Evolutionarily, Thanh Ngọc is closely related to Cymbidium atropalabium, a species with distinct floral morphology, including long and narrow lips adapted to semi-arid environments. If Thanh Ngoc shares a close relationship with this species, it may exhibit similar adaptive traits. Furthermore, Thanh Ngọc is also associated with Cymbidium ensifolium, a widely cultivated Cymbidium species with compact foliage and fragrant flowers. Its proximity to this group suggests that it may share some genetic traits that help it adapt to similar habitats.
– Subgroup I.2: This subgroup includes the following samples: Bach Ngoc Xuan, Tu Thoi, Bach Ngoc Tieu Kieu, Hoang Hoang Sa Pa, MT518381.1 (Cymbidium elegans voucher ORDNA00569) and KC117300.1 (Cymbidium iridioides cultivar Honghoang). In this group, the bootstrap values for this group range from 8 to 42, indicating varying levels of phylogenetic support. The relationship between Bach Ngoc Xuan, Tu Thoi suggests a close connection, possibly belonging to the same Cymbidium lineage or sharing a common origin. Their relatively low genetic divergence suggests they may be variants of the same species or have recently diverged. Bach Ngoc Tieu Kieu and Hoang Hoang Sa Pa showed greater differences, as indicated by a higher bootstrap value (42). This group represents an intermediate branch in Cymbidium evolution, with moderate genetic divergence but still maintaining close relationships. The samples include both wild species (C. elegans, C. iridioides) and cultivated varieties (Hong Hoang Sa pa), making this group significant for understanding the evolutionary history of Cymbidium orchids. They have high potential for conservation and breeding, particularly in developing commercially valuable Cymbidium hybrids.
– Subgroup I.3 were included MF861075.1 (Cymbidium haematodes voucher Z.J.Liu 2503), MK439766.1 (Cymbidium haematodes voucher Z.J.Liu 10160), Cam To, K439771.1 (Cymbidium sinense voucher Z.J.Liu 2596), and MF861068.1 (Cymbidium nanulum voucher Z.J.Liu 2562) and MK439761.1 (Cymbidium cyperifolium voucher Z.J.Liu 3205) samples. Cam To species related closely to Cymbidium haematodes, a species with small, brightly colored flowers found across Asia. This suggests that Cam To may share genetic material with C. haematodes or have a common evolutionary ancestor. It is also closely related to Cymbidium sinense, indicating potential evolutionary overlap or shared ancestry.
Group I.4 included Thanh Truong, Mac Bien and MK439764.1 (Cymbidium goeringii) species based on their positions in the phylogenetic tree, the Thanh Truong species is located within a branch close to a species belonging to the genus Cymbidium, particularly Cymbidium sinense and Cymbidium goeringii. This indicates that Thanh Truong has an evolutionary relationship closely related to the common Asian Cymbidium species. Its proximity to C. sinense and C. goeringii suggests that Thanh Truong may share many genetic traits with these species. It may exhibit similar morphological and biological characteristics, such as flower structure, leaf shape, and habitat preferences.
Mac Bien species is positioned within a branch closely related to Cymbidium ensifolium and Cymbidium kanran. This suggests that Mạc Mac Biên has a close evolutionary relationship with widely distributed Cymbidium species in East Asia. In terms of genetic characteristics, Mạc Biên’s similarity to C. ensifolium and C. kanran suggests that it may possess similar genetic traits, such as cold tolerance or distinct floral morphology. It may also exhibit specific genetic variations, reflecting adaptations to environmental conditions. In summary, the three species as Thanh Truong, Mac Bien and, and MK439764.1 belong to the Cymbidium genus and have close evolutionary relationships with common Asian Cymbidium species. Their proximity suggests they share many genetic and morphological characteristics.
Group II consists of three samples including Kiem Lo Hoi (Cymbidium aloifolium), MF861054.1 (Cymbidium aloifolium voucher Z.J.Liu 6591) and LC714889.1 (Cymbidium aloifolium 2020-RW-21) samples. Based on the phylogenetic tree, Kiem Lo Hoi (Cymbidium aloifolium) forms a distinct branch along with the two samples LC714889.1 and MF861054.1. This has significant implications for the genetic and evolutionary understanding of this sample. Kiem Lo Hoi is most closely related to MF861054.1 (Cymbidium aloifolium voucher Z.J.Liu 6591), indicating that they belong to the same species (Cymbidium aloifolium), but they may represent different strains or originate from different geographical regions. The sample LC714889.1 (Cymbidium aloifolium 2020-RW-21) is also part of this group, forming a branch with a high bootstrap value of 99, which strongly supports the genetic relationships among these samples.
Kiem Lo Hoi and its related samples are clearly distinct from other species within the Cymbidium genus, particularly from the Mac Bien and Thanh Truong groups. This suggests that Cymbidium aloifolium may have diverged early in the evolutionary historical profile of the Cymbidium genus. The branch containing Kiem Lo Hoi has a relatively long length compared to other groups, which may reflect a high mutation rate or accumulated genetic differences. In summary, Kiem Lo Hoi belongs to the Cymbidium aloifolium group, closely related to samples MF861054.1 and LC714889.1, forming a branch with a high bootstrap value (99). This indicates stable genetic relationships within this group. Further research on the genetic differences among strains within Cymbidium aloifolium could provide valuable information for conservation and hybridization efforts.
Group III:
Group III consists of the Dai Mac La cong sample. Dai Mac La cong is positioned at the farthest end of the phylogenetic tree, forming an entirely separate and distinct branch. It has a very large genetic distance from all other samples in the tree, as evidenced by the longest branch length compared to any other sample. The genetic uniqueness of Dai Mac La cong is reflected in the fact that it has no close genetic relatives, indicating that it may have evolved along an entirely independent evolutionary pathway.
Over the course of evolution, Dai Mac La cong may have accumulated numerous unique mutations, making it significantly different from other groups within the Cymbidium genus. Compared to other groups, the closest group to Dai Mac La cong is the Cymbidium aloifolium group (including Kiem Lo Hoi, LC714889.1, and MF861054.1), but there remains a significant genetic distance between them. When compared to groups such as Cymbidium sinense, Cymbidium goeringii, and Cymbidium ensifolium, Dai Mac La cong Cong shows an even greater genetic divergence, suggesting that it may represent a distinct species or a highly mutated lineage within the evolutionary process.Due to its highly distinct genetic characteristics, Dai Mac La cong may contain a unique gene pool, making it crucial for conservation to prevent the loss of rare genetic traits. The significant genetic distance indicates that it could be an ancient lineage or have originated from a separate evolutionary branch within Cymbidium. This could provide valuable insights into the evolutionary historical profile of the genus.
Because of its considerable genetic differences, Dai Mac La cong could serve as an important genetic resource for breeding programs to enhance specific traits in other orchid varieties, particularly in terms of adaptability and floral morphology. In conclusion, Dai Mac La cong is the most genetically distinct sample in the phylogenetic tree, located on an entirely separate branch. It may have diverged early in the evolutionary history of Cymbidium or carried unique mutations that differentiate it from all other species.
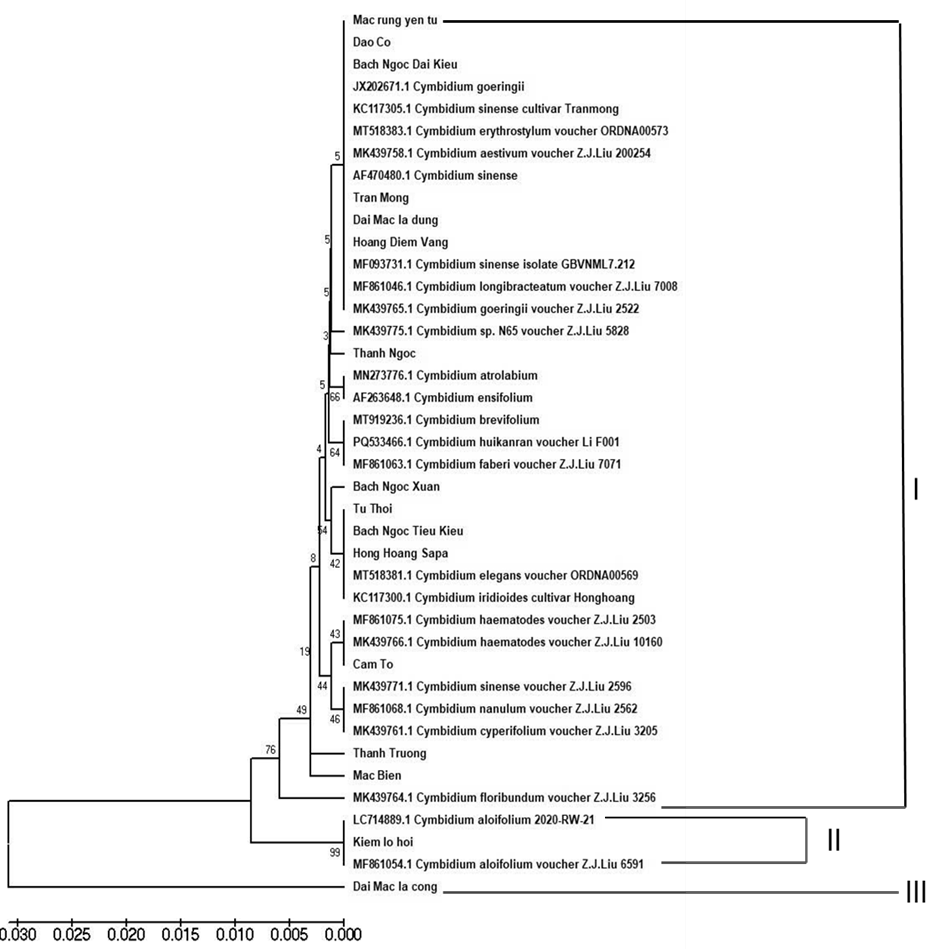
Fig 3. Phylogenetic trees based on matK sequences
Morphology-based methods are generally taken more time- and cost-effective than molecular identification techniques. Nevertheless, to Cymbidium and some land plants, These techniques primarily rely on reproductive structures that are often difficult to distinguish, which limits their accuracy. In the case of leaves and roots, there are few morphological traits that are unique to specific species, making it challenging to differentiate between similar organisms based on these parts alone, often resulting in misidentification. Most of Cymbidium orchid species in Vietnam are now endangered or threatened to extinction due high demand of ornamental trade of these beautiful species. However, it is difficult to distinguish them without their flowers (Fig 5). For example (Dai mac la cong and Dai Bien have similar leaf and some different Cymbidium species). Therefore, it is needed to develop molecular markers to accurately identify these species.

Fig 4: Dai mac la cong and Mac Bien species
CONCLUSION
Our present study is in view to set the first steps in constructing a molecular database of Cymbidium orchids for future assessment of matK gene of Cymbidium species from Vietnam, to contribute to the conservation of these valuable orchids. These results of the matK gene regions in 16 Cymbidium orchid samples showed a successful amplification rate of 70 – 100%. Based on phylogenetic trees, we identified several species within the Cymbidium genus, including Cymbidium sinense (Tran Mong, Dai Mac La Dung, and Hoang Diem Vang), Cymbidium haematodes (Cam To) and Cymbidium aloifolium (Kiem Lo Hoi). However, the matK gene alone has limitations in accurately identifying orchid varieties. Therefore, combining multiple gene regions is recommended, such as ITS + rbcL + matK or ITS + trnH-psbA. This multi-gene approach enhances the accuracy of species identification and genetic relationship analysis within Cymbidium.
Conflict of interest
The authors would like to declare that there have no any conficts of interest in this paper
REFERENCES
- Yang, J.B.; Tang, M.; Li, H.T.; Zhang, Z.R.; Li, D.Z. Complete chloroplast genome of the genus Cymbidium: Lights into the species identification, phylogenetic implications and population genetic analyses. BMC Evol. Biol., 2013, 13, 84.
- Yang, F.X.; Gao, J.; Wei, Y.L.; Ren, R.; Zhang, G.Q.; Lu, C.Q.; Jin, J.P.; Ai, Y.; Wang, Y.Q.; Chen, L.J. The genome of Cymbidium sinense revealed the evolution of orchid traits. Plant Biotechnol. J. 2021, 19, 2501–2516.
- Kim, J.S.; Kim, H.T.; Son, S.-W.; Kim, J.-H. Molecular identification of endangered Korean lady’s slipper orchids (Cypripedium, Orchidaceae) and related taxa. Botany, 2015, 93, 603–610.
- Averyanov, L.; and Averyanova, A.L. Updated checklist of the Orchids of Vietnam. Vietnam National University Publishing House, Hanoi, 2003.
- Li Y.L., Tong Y., Xing F.W. DNA barcoding evaluation and its taxonomic implications in the recently evolved genus Oberonia Lindl. (Orchidaceae) in China. Front. Plant Sci. 2016, 7, 1791. doi: 10.3389/fpls.2016.01791.
- Hoang, N.; Nhung, N.T.; Trang, V.T.H.; Khanh, T.D.; Hung, H.V.Molecular Analysis of Genetic Diversity and Genetic Relationship of Polygonatum kingianum Samples Collected in Northern Mountainous Regions in Vietnam. Eur Chem Bull., 2023, 12(8), 9296-9304.
- Li, X.W.; Yang, Y.; Henry, R.J.; Rossetto, M.; Wang, Y.T.; Chen, S.L. Plant DNA barcoding: From gene to genome. Biol. Rev. Camb. Philos. Soc. 2015, 90, 157–166
- Hebert P.D.N.; Cywinska A.; Ball S.L.; DeWaard J.R. Biological identifications through DNA barcodes. Proc. R. Soc. B Biol. Sci., 2003, 270, 313–321. doi: 10.1098/rspb.2002.2218.
- Kress W.J., Erickson D.L. DNA barcodes: Genes, genomics, and bioinformatics. Proc. Natl. Acad. Sci. USA., 2008, 105, 2761–2762. doi: 10.1073/pnas.0800476105.
- Trung, K.H.; Khanh, T.D.; Ham, L.H.; Duong, T.D.; Khoa, N.T. Molecular phylogeny of the endangered Vietnamese Paphiopedilum species based on the internal transcribed spacer of the nuclear ribosomal DNA. Adv Stu Biol, 2013, 5(7), 337-346. http://dx.doi.org/10.12988/asb.2013.3315.
- Xu, S.; Li, D.; Li, J.; Xiang, X.; Jin, W.; Huang, W.; Jin, X.; Huang, L. Evaluation of the DNA Barcodes in Dendrobium (Orchidaceae) from Mainland Asia. PLoS ONE .,2015, 10, e0115168.
- Singh, H.K.; Parveen, I.; Raghuvanshi, S.; Babbar, S.B. The loci recommended as universal barcodes for plants based on floristic studies may not work with congeneric species as exemplified by DNA barcoding of Dendrobium species. BMC Res., 2012, 5, 42.
- Wu, C.-T.; Gupta, S.K.; Wang, A.Z.-M.; Lo, S.-F.; Kuo, C.; Ko, Y.-J.; Chen, C.; Hsieh, C.-C.; Tsay, H.-S. Internal Transcribed Spacer Sequence Based Identification and Phylogenic Relationship of Herba Dendrobii. J. Food Drug Anal., 2012, 20, 143–151.
- Xu, Q.; Zhang, G.-Q.; Liu, Z.-J.; Luo, Y.-B. Two new species of Dendrobium (Orchidaceae: Epidendroideae) from China: Evidence from morphology and DNA. Phytotaxa., 2014, 174, 15.
- Duong, T.D.; Trung, K.H.; Nghia, L.T.; Thuy, N.T.T.; Hen, N.B.; Khoa, N.T.; Dung, T.H.; Trung, D.M.; Khanh, T.D. Identification of Vietnamese native Dendrobium species based on ribosomal DNA internal transcribed spacer sequence, Adv Stud Biol., 2018, 10(1), 1-12; https://doi.org/10.12988/asb.2018.7823
- Lin, J.-Y.; Lin, B.-Y.; Chang, C.-D.; Liao, S.-C.; Liu, Y.-C.; Wu, W.; Chang, C.-C. Evaluation of chloroplast DNA markers for distinguishing Phalaenopsis species. Sci. Hortic. 2015, 192, 302–310.
- Yukawa, T.; Kinoshita1, A.; Tanaka, N. Molecular Identification Resolves Taxonomic Confusion in Grammatophyllum speciosum Complex (Orchidaceae). Bull. Natl. Mus. Nat. Sci. Ser. B., 2013, 39, 137–145.
- Siripiyasing, P.; Kaenratana, K.; Mokkamuk, P.; Tanne, T.; Sudoon, R.; Chaveerach, A. DNA barcoding of the Cymbidium species (Orchidaceae) in Thailand. Afr. J. Agric. Res., 2012, 7, 393–404.https://doi.org/10.5897/AJAR11.1434
- Khew, G.S.-W.; Chia, T.F. Parentage determination of Vanda Miss Joaquim (Orchidaceae) through two chloroplast genes rbcL and matK. AoB Plants., 2011, plr018.
- Senthilkumar, S.; Xavier, T.F.; Gomathi, G. DNA Barcoding of Vanda Species from the Regions of Shevaroy and Kolli Hills using rbcL gene. Biotechnol. Res. 2017, 3, 126–128.
- Ginibun, F.C.; Saad, M.R.M.; Hong, T.L.; Othman, R.Y.; Khalid, N.; Bhassu, S. Chloroplast DNA Barcoding of Spathoglottis Species for Genetic Conservation. Acta Hortuc. 2010, 878, 453–460
- Xiang, X.G.; Hu, H., Wang,.; DNA barcoding of the recently evolved genus Holcoglossum (Orchidaceae: Aeridinae): a test of DNA barcode candidates. Mol Ecol Res., 2011, 11(6), 1012-1021.https://doi.org/10.1111/j.1755-0998.2011.03044.x.
- De Vere, N.; Rich, T.; Trinder, S.; Long, C. DNA barcoding for plants. Med Mol Biol, 2015, 1245, 101-118. Doi:10.1007/978-1-4939-1966-6_8
- Yao, H.; Song, J.Y.; Ma, X.Y.; Liu, C.; Li, Y.; Xu, H.X.; Han, J.P.; Duan, L.S.; Chen, S.L. Identification of Dendrobium species by a candidate DNA barcode sequence. the chloroplast psbA–trnH intergenic region. Planta Med, 2009, 75 (6),667–669.
- Asahina, H., Shinozaki, J., Masuda, K., Morimitsu, Y., Satake, M. Identification of medicinal Dendrobium species by phylogenetic analyses using matK and rbcL sequences. J. Nat. Med., 2010, 64 (2), 133–138.
- Obara-Okeyo, P.; Kako, S. Genetic diversity and identification of Cymbidium cultivars as measured by random amplified polymorphic DNA (RAPD) markers. Euphytica 1998, 99, 95–101. [Google Scholar] [CrossRef]
- Wang, H.Z.; Wang, Y.D.; Zhou, X.Y.; Ying, Q.C.; Zheng, K.L. Analysis of genetic diversity of 14 species of Cymbidium based on RAPDs and AFLPs. Shi Yan Sheng Wu Xue Bao., 2004, 37, 482–486.
- Wu, Z.X.; Wang, H.Z.; Shi, N.N.; Zhao, Y. The genetic diversity of Cymbidium by ISSR. Yi Chuan., 2008, 30, 627–632.
- Sharma, S.K.; Dkhar, J.; Kumaria, S.; Tandon, P.; Rao, S.R. Assessment of phylogenetic inter-relationships in the genus Cymbidium (Orchidaceae) based on internal transcribed spacer region of rDNA. Gene., 2012, 495, 10–15.
- Moe, K.T.; Zhao, W.; Song, H.S.; Kim, Y.H.; Chung, J.W.; Cho, Y.I.; Park, P.H.; Park, H.S.; Chae, S.C.; Park, Y.J. Development of SSR markers to study diversity in the genus Cymbidium. Biochem. Syst. Ecol., 2010, 38, 585–594.
- Van den Berg, C.; Ryan, A.; Cribb, P.; Chase, M. Molecular phylogenetics of Cymbidium (Orchidaceae: Maxillarieae) sequence data from internal transcribed spacers (ITS) of nuclear ribosomal DNA and plastid matK. Lindleyana., 2002, 17, 102–111.
- Tuong, L.Q.; Tam, T.V.; Dong, D.V.; Duong, T.D.; Cuc, D.T.K.; Thu, P.T.L.;Luong, D.T.; Tuyen, V.T.M.; Giang, N.V.; Tuan, N.T.; Trung, N.T.; Khanh, T.D.; Trung, K.H. Identification of Vietnamese native rose species by using internal transcribed spacer (ITS) sequencing. Plant Cell Biotech Mol Biol., 2020, 11&12, 1-10.
